A new study in Evolution Letters uses experimental evolution to demonstrate how species interactions shape abiotic adaptation, and provides rare insight into the underlying genetics. Lead author James P.J. Hall tells us more.
On the Galapagos island of Daphne Major lives the medium ground finch Geospiza fortis. This small bird eats seeds, large and small, with individuals with larger beaks able to crack open larger seeds. In 1982, a new competitor arrived on the island: big-beaked G. magnirostris, which outcompeted G. fortis for the larger, harder to crack seeds. In response, G. fortis specialised to eat smaller seeds, evolving smaller beaks to do so.
A similar story comes from islands off the coast of Florida, where the small lizard Anolis carolinensis lives in trees, occupying habitats from the ground to the canopy. Introduction of A. sagrei, a related species, displaced A. carolinensis to higher perches in the tree. As anybody who has climbed trees will tell you, the higher you climb, the more unstable and narrow become the branches. Adapting to their new environment, A. carolinensis evolved larger, stickier toepads, helping them to cling on in their more precarious habitat.
In each of these cases, interacting with a competitor altered the relationship that a species has with its environment (its food, or its habitat), generating selective pressure and resulting in evolutionary change. Indeed, one of the fascinating things about evolution is that similar patterns occur in very different species living in very different environments. In the Brockhurst Lab at the University of Sheffield, we investigate evolution in bacteria as a way of understanding general evolutionary processes. Bacteria are excellent study organisms as they’re relatively easy and cheap to grow, and they evolve quickly. Multiple independent populations are allowed to evolve in the lab, allowing us to investigate the repeatability of evolution, and how outcomes vary under different experimental conditions. These experimental populations can be frozen — allowing us to hold ancestral populations in stasis while their descendants evolve — and then thawed, allowing evolved and ancestral bacteria to compete directly against one another. This experimental evolution approach allows us to understand the drivers of evolution in controlled settings. We’re particularly interested in understanding interactions between species: between species of bacteria, and between bacteria and elements such as plasmids and phage.
In one of these evolution experiments, we cultured Pseudomonas fluorescens bacteria in soil microcosms with or without Pseudomonas putida competitors for ~440 generations (read a previous blog post about this experiment here). Both of these bacteria are natural inhabitants of soil, which normally hosts huge, diverse populations of microbes. As part of this experiment we measured the fitness of the evolved P. fluorescens. As expected, all the tested populations had increased in fitness relative to their ancestors, due to adaptation to the experimental culturing regime. However, those which had evolved alongside P. putida did not gain as much fitness, when tested without P. putida, as those which had evolved alone. As with the finches and the lizards, the presence of competitors had affected the evolutionary opportunities available to our bacteria.
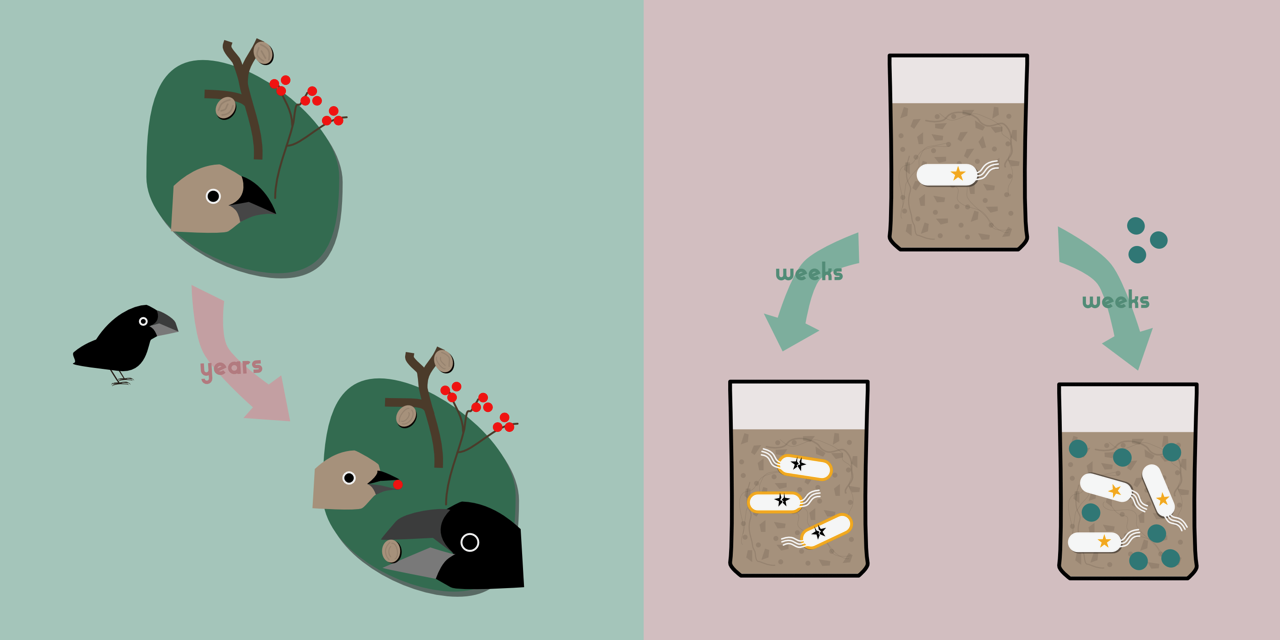
One of the major advantages of studying evolution in bacteria is that sequencing and analysing genomes is usually much easier and cheaper than the equivalent in eukaryotes. These ‘evolve and resequence’ studies can be very powerful at revealing the genetic basis of adaptation. By sequencing our lab-evolved lines and comparing them with the ancestral genomes we can identify mutated loci, and most interestingly, parallel mutations: independent mutations in the same gene occurring in different populations. Parallel mutations are a strong sign of adaptation, and where parallel mutations occur repeatedly in the same treatment (but not in the other) they indicate specific adaptation to those particular conditions. And that is what we saw. Only one gene clearly stood out: the acetate transporter actP. P. fluorescens grown alone almost always had mutations in actP, whereas this mutation was never found in populations co-cultured with P. putida. To investigate these mutants, we grew evolved clones with wild-type actP, or clones with the disrupted actP, in soil microcosms either with or without P. putida competitors. The actP mutants grew faster than the wild-type, but this effect was absent when P. putida was present. We had therefore identified a gene which, when disrupted, enhanced growth in the soil environment. In the presence of a competitor, however, there was no benefit.
ActP’s known activities — nutrient acquisition and toxin sensitivity — make intuitive sense in the context of between-species interactions. Competitors change the local environment by altering nutrient availability and neutralising common toxins, which is likely to have implications for the costs and benefits of maintaining a functioning ActP. We also identified two cases where a broken transporter from the same family as ActP was reactivated by mutations reverting a premature stop codon. Both of these were in clones which had evolved alongside P. putida, hinting at a generally important role of nutrient transporters in species interactions. While for the lizards and the finches competition resulted in the evolution of specialism, it seems that for our soil-dwelling bacteria the presence of a competitor selected individuals that had a broader range of ecological functions.
Our work provides a rare glimpse of genetics underlying the evolution of species interactions. Future experiments aim to get to the bottom of ActP’s role in adaptation, and its costs and benefits in interactions with species other than P. putida. It would also be interesting to see how readily actP mutants revert when single-species-evolved P. fluorescens is introduced to P. putida, and to investigate the function of the re-activated transporter. From finches to lizards to soil bacteria, species interactions play a key role in evolution, and evolution experiments offer a powerful opportunity to explore their effects.
James P. J. Hall (@jpjhall) is a postdoctoral research assistant at the University of Sheffield, Department of Animal & Plant Sciences. The original study is freely available to read and download from Evolution Letters here.

