A new study in Evolution Letters experimentally investigated how population dynamics in colonising populations influence the evolution of infectious disease. Here, lead author Louise Nørgaard describes the experimental approaches that led to the group’s interesting findings.
Infectious disease is a major threat to human and wildlife well-being and epidemiological theory helps us understand how a pathogen is expected to spread through a population. Most host-pathogen studies, however, have made the simplifying assumption that the spread and evolution of disease occurs in stable and nondynamic host populations. Wild host populations are instead ever changing and dynamic, with population growth rates and sizes fluctuating across time and space, as is seen when species expand their habitat range, or in populations with high extinction and colonization rates. This limitation in our understanding of the mechanism in play, when a pathogen is confronted with colonizing host populations, was the major motivation behind our study. This led us to perform a range of laboratory experiments to explore the influence of host population dynamics on the performance of an invading pathogen at different stages of the invasion process, from the initial entry of an infected carrier into a population to the subsequent spread of new carriers to populations elsewhere.
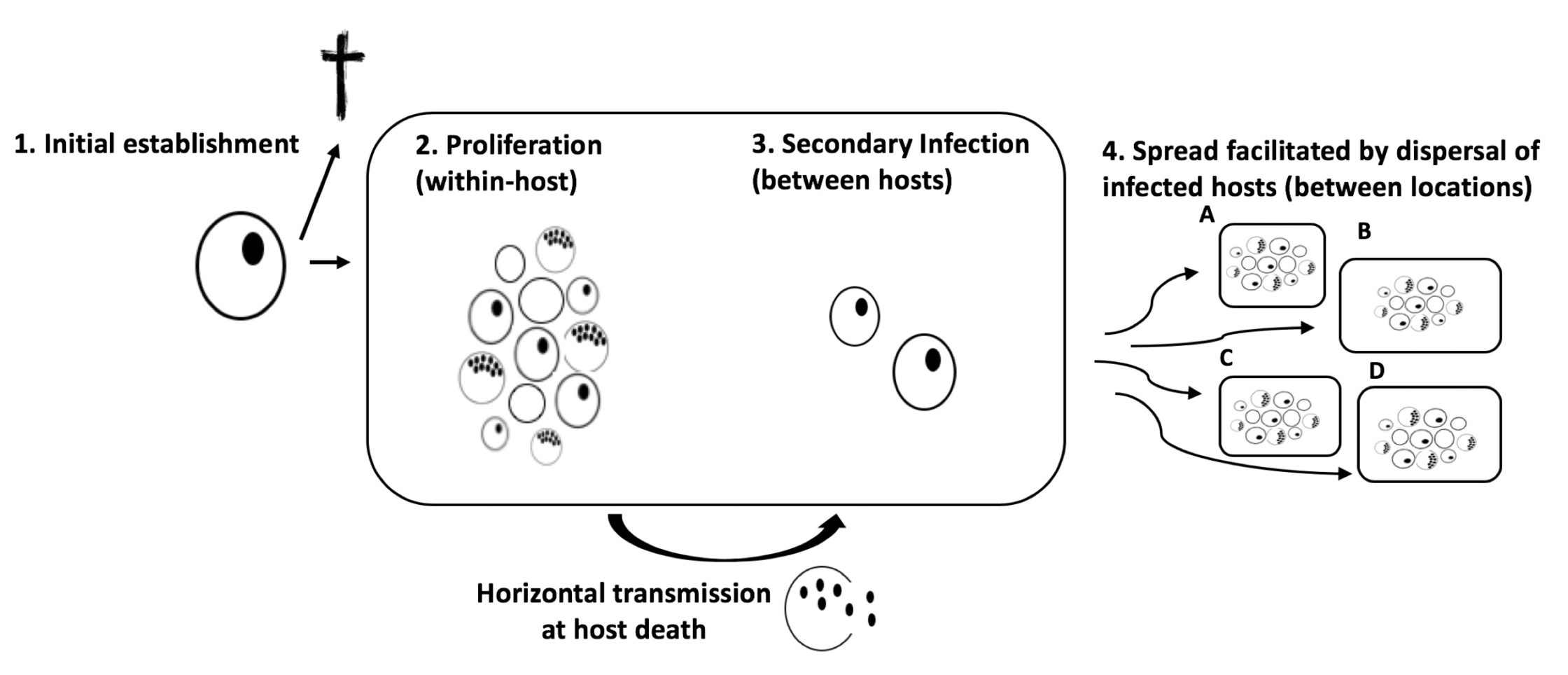
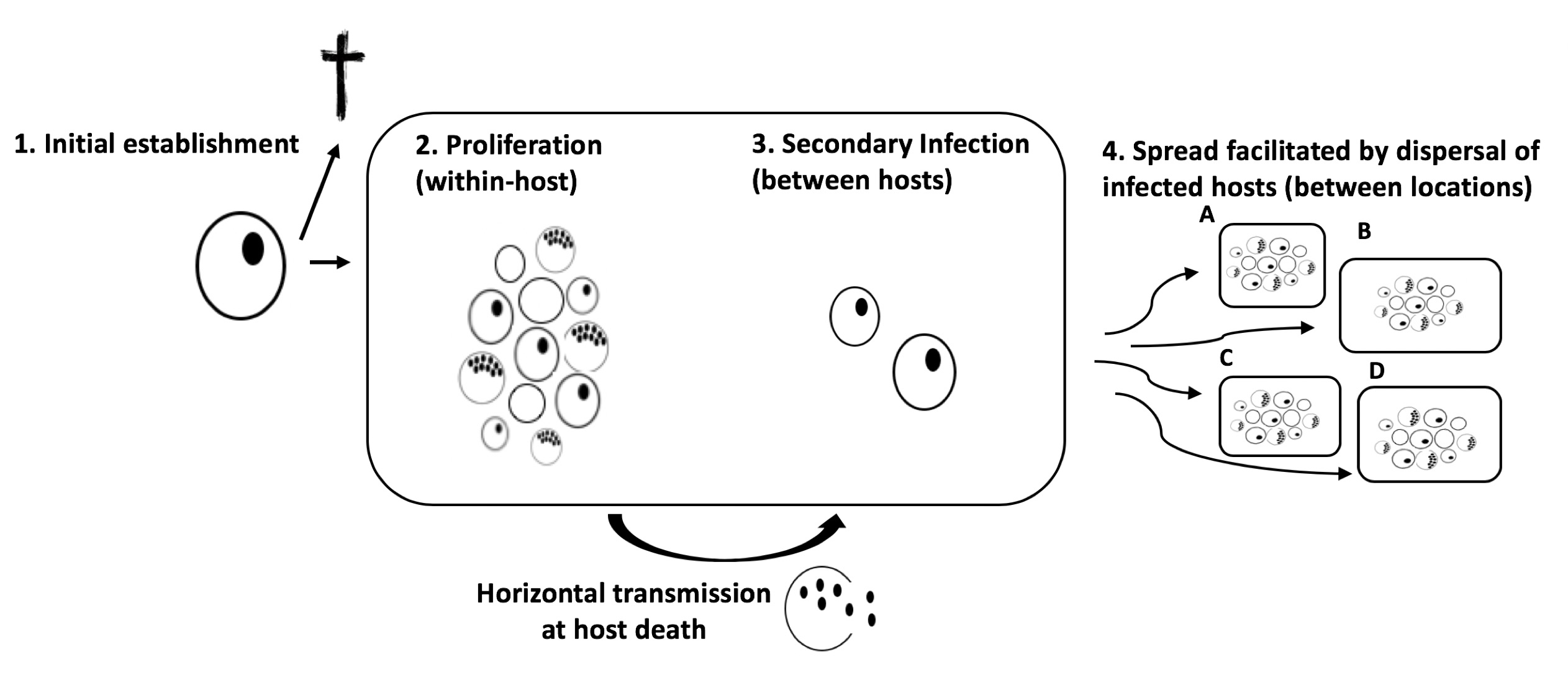
To do this, we needed a system in which we could control a range of environmental, demographic, and disease related variables. For this purpose, the water flea Daphnia magna and its bacterial gram-positive pathogen, Pasteuria ramosa, appeared to be particularly useful. In this system, the clonally reproducing host becomes infected upon filter-feeding, when spores are present in the water. The spores are then activated inside the host, start to proliferate and fill up the host body cavity, and eventually kill the host. The spores are then released from the dying cadaver, and a new infection cycle can be initiated. These dynamics allowed us to control for genetic background in the host, but also to tease apart the four parasite invasion steps, ranging from 1) initial invasion of an infected carrier, 2) proliferation and production of mature transmission spores within the infected host, 3) spread to new susceptible hosts (secondary infection) within the same population, and finally, 4) the parasite must spread between populations to infect hosts in a patchy landscape.
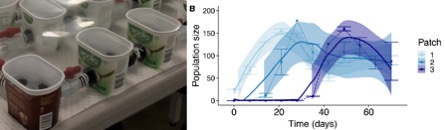
With an experimental system in hand, we proceeded to first explore how colonization dynamics in Daphnia populations arise as it spreads through a patchy landscape. We seeded experimental patches that were connected to a series of new and uninhabited patches and monitored the growth in population size over time in each patch. We found characteristic population dynamics consisting of an initial phase of rapid growth followed by a short phase of overshoot, before finally reaching a stable carrying capacity. Importantly, those population dynamics were very distinct and repeatable between habitat patches, allowing us to test disease dynamics in each of the three host colonization phases.
In the next experiment, we tested the first two steps of the pathogen infection process; initial establishment and proliferation capacity when confronted with each of the three characteristic host colonization phases (rapid growth, overshoot, and stationary phase). Here, we found that pathogens generally established with higher success in host populations that had just colonized a new habitat patch, but the five different pathogen genotypes tested differed in their success rates. Then we then quantified the spore production in pathogens that successfully established, and again found that most pathogens had highest spore production in recently established populations, but with clear pathogen specific responses.
This got us excited, and we moved on to explore the influence of infection on dispersal behavior, and to understand whether Daphnia, like most humans, prefer to stay at home and rest when sick. We found that the behavioral response to infection was highly dependent on parasite genotype; some parasites reduced dispersal rate of their host significantly, others barely influenced the rate at which individuals moved through space. Most importantly however, the driving factor in determining how far an infected carrier moved appeared to be the lifespan; infection reduced lifespan, and the pathogen genotypes differed markedly in how quickly they killed their host and how much they impacted on dispersal.
Overall, our results show that pathogen invasion fitness will be highest in newly colonized host patches, but not all pathogens will have the dispersal capacity to routinely exploit these transient conditions. Our results suggested that pathogen traits optimizing pathogen fitness in a growing population may not be the same in a host population at carrying capacity, and we identified a complex array of successful pathogen strategies, depending on the demographic conditions of the host population. This clearly demonstrated the potential for different pathogen strategies or genotypes to be maintained across a landscape in which host population vary in their colonization history or demography.
Louise Nørgaard is a PhD candidate in Dr Matt Hall’s lab at the School of Biological Sciences, Monash University. The original study is freely available to read and download at Evolution Letters.