In our new author blog, Joel McGlothlin explains how natural selection and genetic constraint interact to shape adaptive radiations.
Ever since the Modern Synthesis of the early 20th century, biologists have had a pretty good understanding of how evolution works. Mutation generates genetic variation in a population, and natural selection sorts through it, keeping things that fit the current environment and weeding out things that don’t. Along the way, populations sometimes split into two, adapting to different environments and becoming new species in the process.
All evolutionary biologists agree about these basics of the evolutionary process, but there is still a lot of controversy about the relative importance of its different aspects. For example, some scientists place more weight on the importance of natural selection as an explanation for biological diversity, while others think that genetic variation plays a guiding role in constraining which paths adaptation can take.
It was with this big question in mind that my colleagues and I started on a large-scale quantitative genetic study of Anolis lizards. Anoles are icons of what is known as adaptive radiation, an evolutionary pattern involving the origin of group of related species that differ in both their appearance and ecological role. In our paper, published today in Evolution Letters, we describe our efforts to disentangle the roles of natural selection and genetic constraints in generating this diversity of form.

My co-authors (Butch Brodie, Jonathan Losos, Megan Kobiela, Helen Wright, Luke Mahler, and Jason Kolbe) and I estimated patterns of genetic variation underlying ecologically important traits (called “G” for short) in seven different species of anoles. We captured hundreds of adult lizards from three islands (Puerto Rico, Jamaica, and South Bimini, Bahamas) and returned them to the lab, where we bred thousands of babies over the course of several years. The breeding effort was massive and was only possible with the help of a huge team of students and technicians working everyday in the animal facility. Every one of these baby lizards was X‑rayed so we could measure skeletal traits like head shape and limb length to generate an estimate of for G for each species.
These measurements of G allowed us to test a number of hypotheses about genetic variation and its influence on the evolutionary process. First, we asked whether the evolutionary differentiation of anole morphology was related to genetic variation described by G. A classic prediction in evolutionary quantitative genetics is that species should diverge along the “genetic line of least of resistance,” or “gmax,‚” defined as the combination of traits with the most genetic variation, for at least a few million years. In anoles, we found that divergence of morphology was indeed biased in the direction of gmax, even though the species had been separated for up to 40 million years.
This result was surprising to us, because the relationship between gmax and divergence is predicted to decay over time. One reason for this decay is that G itself is expected to change as species evolve, which might obscure the relationship by changing gmax. Because we measured G in multiple species, we were able to measure this change. Our results show that G changes significantly in size, shape, and orientation. Why, then, did the relationship between G and divergence persist?
Using a method called genetic covariance tensor analysis, we broke change in G down into its component parts. This showed us that although G changed, the way it changed did not alter the genetic lines of least resistance. In fact, most of the change in G involved growing and shrinking in the direction of gmax.
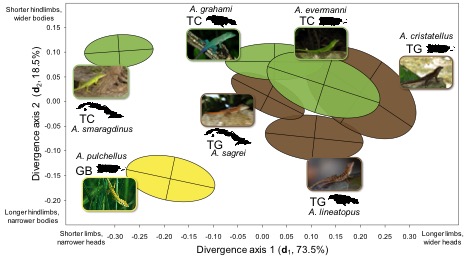
Putting all these results together, we found that three axes—genetic variation within species (gmax), change in morphology across species, and change in genetic variation across species—were all aligned with one another. This result is open to a wide variety of interpretations, which we discuss further in the paper. At one extreme, genetic constraint could be the whole story. Species are constrained to evolve along genetic lines of least resistance, and even though these constraints evolve, the constraints themselves are constrained to evolve in certain directions. At the other extreme, selection reigns supreme, and divergence resembles gmax because they have both been shaped by adaptation. The reality is probably somewhere in between.
When we began this study, we thought we might be able to show that either selection or genetic constraint determined the paths of adaptive radiation, but instead, we may have demonstrated just how difficult these forces are to separate. At least in anoles, constraint shapes the evolutionary response to selection, but also evolves in response to selection in such a way to keep the two entwined. Perhaps it’s this never-ending creative dance that makes evolution so interesting in the first place.
Joel McGlothlin is Assistant Professor in the Department of Biological Sciences, Virginia Tech. The study is freely available to read and download here.

