A new study published in Evolution Letters demonstrates that rapid adaptation in natural populations of sticklebacks results from both genetic linkage (where multiple mutations located close together in the genome are inherited together) and pleiotropy (where the same mutation leads to changes in many traits). Lead author Dr Sophie Archambeault tells us more.
When organisms enter new environments, they are faced with many environmental challenges such as changes in temperature, salinity, predators, prey and parasites. These environmental changes impose new selection pressures on the organisms and can drive adaptive evolution. Evolutionary biologists like myself are interested in understanding the links between these selection pressures, the adaptive changes, and the mutations that cause these changes. Making these connections is critical for understanding the formation of the diversity of life present today and for predicting the evolutionary trajectories of future populations in response to changing environments.
The growth of genomic resources over the last two decades has improved our ability to identify the genomic regions associated with adaptive changes (i.e., “map the traits”) in natural populations, including butterflies, petunias, deer mice, and stickleback fish. Efforts to map traits observed in natural populations have uncovered a surprising recurring pattern. Some regions of the genome control more traits than expected by chance. In other words, traits often cluster in the genome. This finding is consistent with either of two phenomena: pleiotropy or linkage.
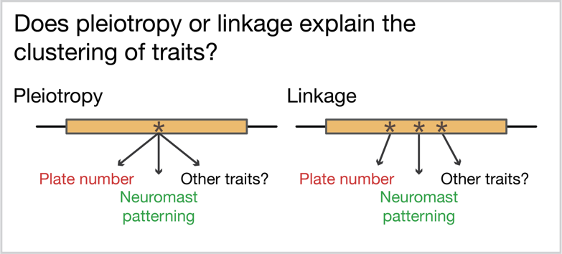
The question we set out to answer: is clustering of traits driven by pleiotropy or linkage of causative mutations?
Pleiotropy refers to a single mutation in the DNA that affects more than one trait. This is often observed in diseases caused by mutations in developmental genes because these genes are often critical for the correct formation of multiple tissues. Linkage describes a different situation, where multiple mutations each affecting a single trait are nonetheless inherited together because the mutations are physically located near each other in the genome. Pleiotropy is thought to constrain adaptation because multiple trait changes are unlikely to all be favorable in the new environment and the changes cannot be selected separately by selection. In contrast, linkage is predicted to be favored during adaptation because each trait can be “screened” individually by selection, and linkage ensures that traits favorable in the new environment are inherited together. This may be particularly important if organisms still occasionally breed with the organisms adapted to the old environment. However, very few instances of clustered traits have been dissected to the level of the mutation, meaning we really don’t know whether linkage or pleiotropy is more common!
We therefore set about to figure out whether linkage or pleiotropy explains the clustering of traits observed in stickleback fish, an important model species for studying adaptation in vertebrates. Marine sticklebacks have adapted to freshwater environments thousands of times across the Northern hemisphere since the last ice age (~12,000 years ago). This repeated adaptation to freshwater has led to repeated changes in traits such as reduction of bony elements (opercula, spines, and plates) and patterning of the individual sensory organs (neuromasts) that detect water movement. Most of the variation in bony plate number and neuromast patterning is controlled by the developmental gene Eda. Nearly all sticklebacks with reduced plate numbers share a stretch of 16,000 DNA base pairs on chromosome 4. We wanted to find the individual mutation (or mutations) within this genomic region that controls bony plate number and determine whether it also affects neuromast patterning and other traits that have previously mapped to chromosome 4.
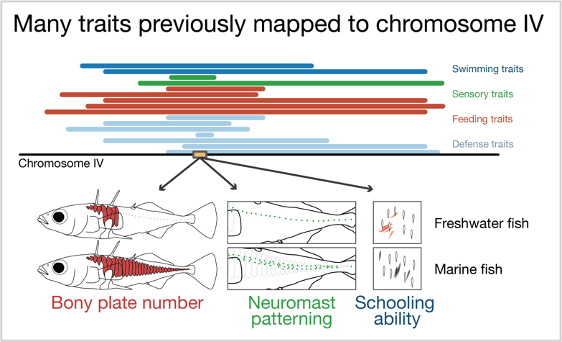
Previous work has found that many traits map to chromosome 4 in the stickleback genome. These include swimming, sensory, feeding and defense traits. Three traits map to a developmental gene on chromosome 4, Eda (yellow box). These traits are number of bony plates, patterning of the mechanosensory organs (neuromasts), and the ability of fish to school with a model school of fish!
The first challenge was finding fish that possessed both freshwater and marine mutations within this small genomic region. We lucked out on finding the rare carriers of marine and freshwater alleles in both marine and lake populations of sticklebacks near our old lab in Seattle (Figures 3 & 4). The second challenge was packing up and moving the lab (including thousands of live fish) to our new home in Switzerland while madly performing thousands of PCRs before shipping all the boxes of frozen DNA! Once the fish and DNA were safely shipped to our new laboratory home in Switzerland, it was time to gather the data. Our final challenge was organizing the immense amount of data collection – up to 91 different traits per fish for almost 1500 fish! Fortunately, we assembled a great phenotyping team, including undergraduate students Luis Bärtschi and Aurélie Merminod!
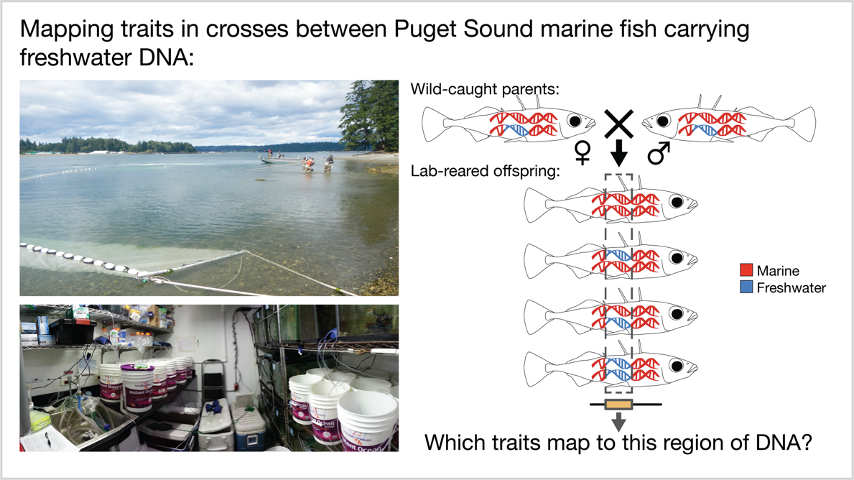
Marine fish were caught in a beach seine in Puget Sound in the summer of 2016 with help from the Washington Department of Fish and Wildlife. Almost 900 fish were transported to the lab and individually genotyped to identify the rare marine carriers of freshwater DNA at Eda. These rare marine fish were then crossed to each other to make 11 separate crosses that differed only in the presence and length of the stretch of freshwater DNA.
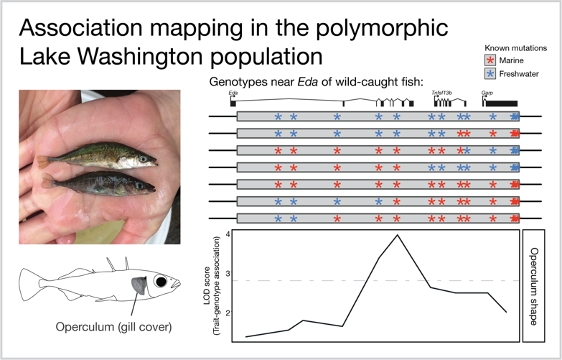
Freshwater fish were caught from a polymorphic population in Lake Washington, Seattle. A female (top) and male (bottom) display mating color differences. Within the 16,000 base pair stretch of DNA surrounding the gene Eda, a subset of these fish carried a mix of known marine and freshwater mutations. This mix of mutations allowed us to compare the association between each mutation and each trait to find the mutation with the strongest association. Depicted here is the measurement of operculum shape, which is most strongly associated with a mutation near the middle of the region.
So, is it linkage or pleiotropy? We found strong evidence that both are at work within this small region of the genome. We found that a 1,400 base pair region within the first intron of Eda has large, pleiotropic effects on plate number, neuromast patterning and neuromast number. But we also found that other regions had small effects on operculum shape, plate number and neuromast patterning. Together these findings suggest that plate number and neuromast patterning is largely controlled by a small region of intron 1 (likely pleiotropy!) but different nearby mutations more broadly explain the clustering of traits on chromosome IV (linkage!). We are continuing to focus on the 1,400 base pair region – it contains 19 candidate mutations, which are being functionally tested for pleiotropic effects on plate number and neuromast patterning in Katie Peichel’s lab in Switzerland.
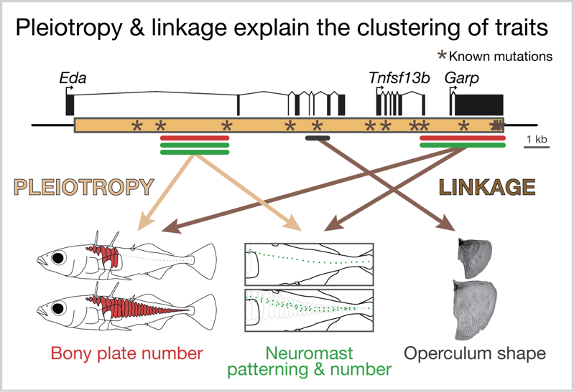
Both pleiotropy and linkage likely explain the clustering of the traits surrounding Eda.
What do these results mean for organisms adapting to new environments? First, mutations in or near developmental genes like Eda are likely to affect multiple traits, highlighting constraints on adaptation via developmental genes. Second, pleiotropy weakens the correlation between repeated trait evolution and adaption – traits may evolve due to selection for correlated traits. For example, if neuromast patterning and plate number are controlled by the same mutation, we need additional information to know which trait is adaptive! Similarly, other mutations may be more likely to contribute to trait evolution when they are physically linked to genes that play large roles in adaptation. Finally, linkage of multiple mutations does appear to be adaptive. In this example, selection has maintained linkage within a 16,000 base pair region, when only 1,400 base pairs are required for the large shifts in plate number and neuromast patterning. Perhaps the linkage of multiple mutations controlling known and as-of-yet unknown traits helps explain the surprising speed with which stickleback can adapt to freshwater. It will be exciting to study the adaptive process while it is happening in sticklebacks and other systems to understand the general prevalence and importance of pleiotropy and linkage during adaptation.
Dr Sophie Archambeault is a postdoctoral scholar at UC Berkeley. The original article is freely available to read and download from Evolution Letters.


