A new study published in Evolution Letters suggests that large blocks of linked genes play a key role in maintaining adaptation to local conditions despite extensive connectivity across latitudes in an estuarine fish. Here, Nina Overgaard Therkildsen and her co-authors explain how this study highlights that field observations of phenotypic similarity across a species’ range can deceptively mask remarkable levels of genetic divergence.
Darker coat color in mice inhabiting patches of dark substrate for better camouflage, crickets that evolve to be silent when confronted with parasites attracted by their calls, and stickleback fish that consistently lose body armor when colonizing freshwater environments that harbor completely different threats than their ancestral ocean environment. These are just a few well-documented examples where adaptation to contrasting local conditions across a species range is highly conspicuous and can readily be observed in the field. In many other cases, such adaptive divergence can be hidden from plain sight. The Atlantic silverside (Menidia menidia) represents a case in point.
A small estuarine forage fish, the Atlantic silverside is abundant along the entire east coast of North America from northern Florida to the Gulf of St Lawrence, spanning one of the steepest temperature gradients in the world. The contrast between the subtropical marshes in the southern end of the distribution range and the chilly temperate waters inhabited in the north results in a nearly threefold difference in the length of the growing season for Atlantic silversides across latitudes. We might expect that such vast variation in temperature and seasonality should substantially affect the growth and life history of any fish (whose body temperature depends on the environment). Yet, in the wild, Atlantic silversides have a fixed one-year life span and grow to roughly the same size across their entire distribution range. Field observations thus do not indicate notable spatial patterns of differentiation.

Atlantic silversides schooling in their natural habitat (photo credit: Jakob Snyder)
It was only when David Conover, a former Professor at Stony Brook University and co-author on the new paper, along with his team started raising silversides from different latitudes in the lab under common conditions that the remarkable degree of local adaptation exhibited by this species became apparent. It turned out that to compensate for the short growing season, silversides inhabiting northern habitats have evolved much faster growth rates, while tradeoffs with predator avoidance have kept growth rates lower in the south.
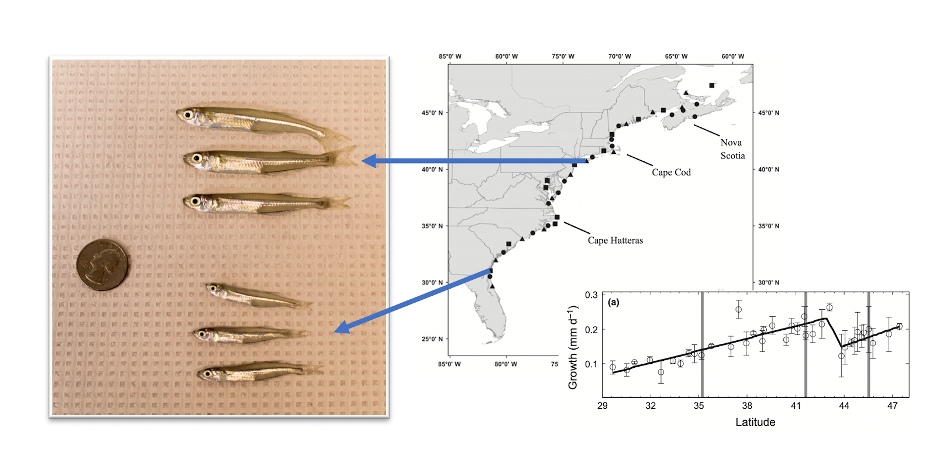
Left: Photo illustrating the difference in growth rates between Atlantic silversides with parents from New York (top) vs. parents from Georgia (bottom) when raised under common conditions in the lab. All fish are of identical age (photo credit: Nina Overgaard Therkildsen and Maria Akopyan). Right: Growth rates estimated in common garden experiments on Atlantic silversides collected from 39 different locations across the latitudinal range (results from Hice et al. 2012. Ecology Letters 15: 568–575).
As a result, the faster intrinsic growth capacity in the north almost exactly counteracts the environmental effect of colder temperatures on growth, so that fish in the wild all get to about the same size. This pattern – genetic influences on a trait counteracting environmental influences – is called countergradient variation and can be very deceptive. This is because field observations give the impression that all fish are the same, but the phenotypic similarity is, in fact, maintained by a surprising degree of functional genetic divergence between populations. Decades of common garden and field studies by Conover and his team have revealed that in addition to growth rates, a whole suite of other fitness-related traits including morphology, physiology, and behavior show similar cryptic countergradient genetic variation in the Atlantic silversides – variation that was invisible until revealed with the right approach.
Now, our new study unveils another level of previously invisible divergence in this species, this time directly in the DNA. Earlier molecular studies using allozomes or mitochondrial DNA had suggested very limited genetic differentiation with only weak structuring on broad regional scales. Our scan of nearly 2 million genetic variants confirmed that the majority of the genome shows minimal differentiation along the coastline. Yet, against this backdrop we find massive chromosomal blocks that contain hundreds of genes for which virtually all southern fish hold variants that are rarely found in the northern part of the range. That is to say, southern and northern populations each harbor blocks of unique genetic variants that are not shared, even though signals in the rest of the genome suggest that fish frequently intermix up and down the coast (and this is backed up by microchemistry-based dispersal tracking). The large genetic differences are likely maintained by natural selection against the southern blocks of genes in northern locations and vice versa.
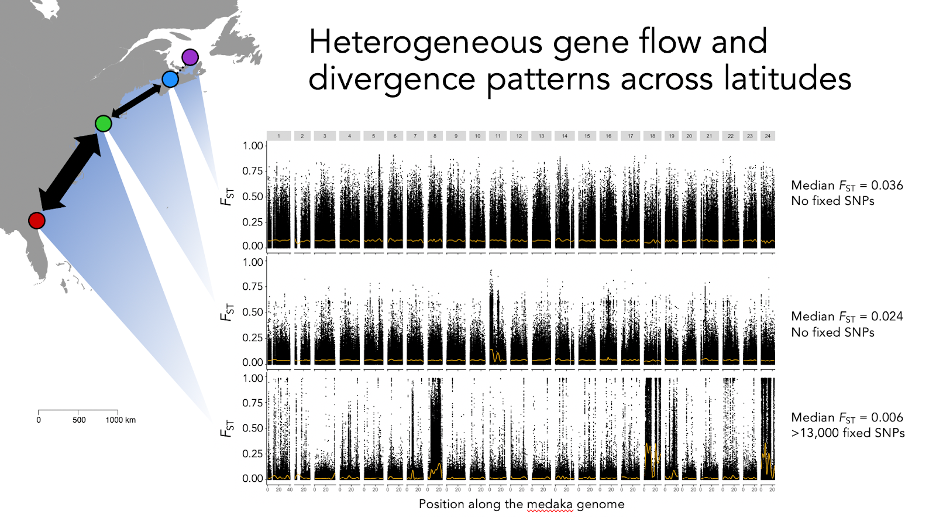
Plots of the level of genetic differentiation (FST) for each genetic variant (ordered along the genome) in different pairwise comparisons of sampling locations. We see that the baseline level of differentiation is highest among the two northernmost locations (but still relatively low as evident from the colored sliding mean lines) and close to zero across the southern stretch of the distribution range. Strikingly, we see that the minimal genome-wide differentiation along the southern coastline is interspersed with large blocks of extreme differentiation on multiple chromosomes.
While we don’t yet know the functional significance of the highly differentiated blocks, they contain genes related to traits known to differ across latitudes in Atlantic silversides, including lipid storage, metabolic rate, and spawning behavior, further supporting their involvement in the well-described local adaptation. Especially intriguing is the tight clustering of the highly differentiated genes into large blocks that extend across much of four different chromosomes but do not seem to recombine, indicating that they may represent chromosomal inversions. The advantage of this genome structure may be that offspring inherit all the southern or all the northern variants of these genes in one package – setting them up to have the right combination without mixing of southern and northern adaptations. In this view, the architecture of the genome itself is an adaptation that promotes adaptability.
As we can now screen variation across the entire genome of virtually any organism, similar inversions are being discovered in an increasing number of species. Yet, while these inversions tend to show elevated levels of differentiation compared to the rest of the genome, we have only seen a few other examples of near-complete fixation of massive blocks of genes across populations with minimal genome-wide differentiation. The Atlantic silverside thus represents an important opportunity for further exploration of what role inversions and other aspects of genomic architecture play in enabling local adaptation in the face of high gene flow and how selection can maintain such extreme levels of genomic differentiation in a species that at first glance seemed deceptively homogeneous across its distribution range.
In combination, common garden studies and population genomic analysis have provided an extraordinary view into intraspecific variation that was invisible to us until we used the right tools to explore it. Given that countergradient variation is now known to be common in nature, the Atlantic silverside serves as an important cautionary tale about assuming homogeneity based on field observations alone. The underlying genetic variation may be critical for conserving evolutionary potential in the face of human impact, as illustrated when Atlantic silversides were subjected to strong fisheries pressure. In fact, the remarkable adaptability of this little fish may serve as an important model in adaptability to future climate change.
Nina Overgaard Therkildsen is an Assistant Professor in the Department of Natural Resources at Cornell University and conducted the study in collaboration with Aryn Wilder who is a Researcher at the San Diego Zoo Institute for Conservation Research, Steve Palumbi who is a Professor at Stanford University’s Hopkins Marine Station and David Conover who is an Emeritus Professor at the University of Oregon.
The original study is freely available to read and download from Evolution Letters.